Acetylserotonin O-methyltransferase
| ASMT | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | ASMT, ASMTY, HIOMT, HIOMTY, acetylserotonin O-methyltransferase | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 402500, 300015; MGI: 96090; HomoloGene: 48261; GeneCards: ASMT; OMA:ASMT - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| acetylserotonin O-methyltransferase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 2.1.1.4 | ||||||||
| CAS no. | 9029-77-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
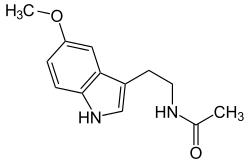
N-Acetylserotonin O-methyltransferase, also known as ASMT, is an enzyme which catalyzes the final reaction in melatonin biosynthesis: converting Normelatonin to melatonin. This reaction is embedded in the more general tryptophan metabolism pathway. The enzyme also catalyzes a second reaction in tryptophan metabolism: the conversion of 5-hydroxy-indoleacetate to 5-methoxy-indoleacetate. The other enzyme which catalyzes this reaction is N-acetylserotonin-o-methyltransferase-like-protein.[5]
Synonyms
[edit]Synonyms of N- Acetylserotonin O-methyltransferase are Hydroxyindole O-methyltransferase (HIOMT), Acetylserotonin O-methyltransferase (ASMT), Acetylserotonin N-methyltransferase, Acetylserotonin methyltransferase (Y chromosome).[6] The most commonly used synonym is Hydroxyindole O-methyltransferase (HIOMT).
Species distribution
[edit]N- Acetylserotonin O-methyltransferase is found in both prokaryotes and eukaryotes. It is found in the bacteria Rhodopirellula baltica and Chromobacterium violaceum. It is also found in the following eukaryotes: Gallus gallus (chicken), Bos taurus (cow), Homo sapiens (human), Macaca mulatta (rhesus monkey), and Rattus norvegicus (rat).[6]
Tissue distribution
[edit]Recent studies found messenger RNA (mRNA) transcripts of the HOIMT gene in B lymphocytes, T helper lymphocytes, cytoxic T lymphocytes, and natural killer lymphocytes in humans. This finding, in conjunction with research on alternative splicing of the HOIMT hnRNA, suggests that Hydroxyindole O-methyltransferase (synonym for N- Acetylserotonin O-methyltransferase) plays a role in the human immune system, in addition to its endocrine and nervous system functions. In other words, the gene may be expressed in various isoforms in different cells of the body.[7]
Gene
[edit]N-Acetylserotonin O-methyltransferase is an enzyme that is coded for by genes located on the pseudoautosomal region of the X and Y chromosome, and is most abundantly found in the pineal gland and retina of humans.[8]
The human HOIMT gene is approximately 35 kb in length and contains 9-10 exons. The gene can be alternatively spliced to form at least three possible isoforms, although each of these isoforms has the same role in the biosynthesis of melatonin. It has also been found that the gene contains multiple promoter regions, an indication that multiple mechanisms of regulation exist.[9]
In humans the ASMT enzyme is encoded by the pseudoautosomal ASMT gene. A copy exists near the endcaps of the short arms of both the X chromosome and the Y chromosome.[10][9]
Structure
[edit]N-Acetylserotonin O-methyltransferase (ASMT) adopts the canonical SAM-dependent methyltransferase architecture in its C-terminal catalytic domain, where a seven-stranded β-sheet flanked by α-helices forms the S-adenosyl-L-methionine (SAM) binding pocket and catalytic site characteristic of class I (Rossmann-like) methyltransferases. ASMT functions as a homodimer, with the predominantly helical N-terminal domains of two monomers intertwining to stabilize the quaternary structure and contribute to proper active-site geometry. In the human X-ray structure, the substrate-binding pocket positions the 5-hydroxy group of N-acetylserotonin for nucleophilic attack on the SAM-derived methyl group, while conserved residues coordinate SAM or S-adenosyl-L-homocysteine (SAH) and interact with the indole ring to enforce regioselective O-methyl transfer.[11]
The structure of N- Acetylserotonin O-methyltransferase has been determined by X-ray diffraction.[12]
Function
[edit]N-Acetylserotonin O-methyltransferase can be classified under three types of enzyme functional groups: transferases, one-carbon group transferrers, and methyltransferases.[6]
It catalyzes two reactions in the tryptophan metabolism pathway, and both can be traced back to serotonin. Serotonin has many fates in this pathway, and N- Acetylserotonin O-methyltransferase catalyzes reactions in two of these fates. The enzyme has been studied most for its catalysis of the final step of the pathway from serotonin to melatonin, but it also catalyzes one of the reactions in the many step process of serotonin → 5-Methoxy-indolacetate.
Reactions catalyzed
[edit]In one metabolic pathway from tryptophan, N-Acetylserotonin O-methyltransferase catalyzes two separate reactions. The first is the reaction of N-acetylserotonin to melatonin. S-adenosyl-L-methionine (SAM) is used as the source of the methyl group and is converted to S-adenosyl-L-homocysteine (SAH).[6][13]
The second reaction catalyzed by the enzyme is later in the pathway, after serotonin has been metabolised to 5-hydroxyindoleacetic acid. This is further converted to 5-methoxyindoleacetate, with the same SAM cofactor.[6][14]
Clinical significance
[edit]Cancer
[edit]There is evidence of high HIOMT gene expression in pineal parenchymal tumors (PPTs). This finding has led to the study of varying gene expression as a diagnostic marker for such tumors. Abnormally high levels of HIOMT in these glands could serve as an indication of the existence of PPTs in the brain.[15]
Psychiatric disorders
[edit]Melatonin levels are used as a trait marker for mood disorders, meaning that abnormal levels of melatonin can be used in conjunction with other diagnostic criteria to determine whether a mood disorder (e.g. Seasonal affective disorder, bipolar disorder, or major depressive disorder) exists. Melatonin levels can also be used as a state marker, contributing to conclusions on the severity of a patient's illness at a given point in time. Because studies have shown a direct correlation between the amount of hydroxyindole-O-methyltransferase in the pineal gland and the melatonin level, additional knowledge of HIOMT could provide valuable insight on the nature and onset of these impairing disorders.[16]
Developmental disorders
[edit]Subjects with autism were found to have significantly lower levels of melatonin and acetylserotonin O-methyltransferase (ASMT) than controls.[17][18]
Linkage analysis
[edit]High frequency polymorphism exists on the PAR region of the sex chromosomes, where the HIOMT gene is located. Linkage analysis of a diseased locus with high frequency polymorphism of this region could lead to vital information about the role of this gene in genetic disorders.[19]
Research
[edit]HIOMT as the limiting reagent in the melatonin biosynthetic pathway
There has been some controversy over the regulatory power of hydroxyindole-O-methyltransferase in the production of melatonin. In 2001, it was argued that another enzyme in the pathway, N-acetyl transferase (NAT) was the limiting reagent in the production of melatonin.[20] Recent findings, however, have suggested that HIOMT, not NAT, is the limiting reagent, and a direct correlation between HIOMT expression and melatonin levels has been shown to exist.[21]
See also
[edit]References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000196433 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000093806 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, et al. (January 2006). "From genomics to chemical genomics: new developments in KEGG". Nucleic Acids Research. 34 (Database issue): D354 – D357. doi:10.1093/nar/gkj102. PMC 1347464. PMID 16381885. [See also comments in Thomson's website]
- ^ a b c d e Enzyme 2.1.1.4 at KEGG Pathway Database.
- ^ Pozo D, García-Mauriño S, Guerrero JM, Calvo JR (August 2004). "mRNA expression of nuclear receptor RZR/RORalpha, melatonin membrane receptor MT, and hydroxindole-O-methyltransferase in different populations of human immune cells". Journal of Pineal Research. 37 (1): 48–54. doi:10.1111/j.1600-079X.2004.00135.x. PMID 15230868. S2CID 22197004.
- ^ Online Mendelian Inheritance in Man (OMIM): x-chromosomal ASMT - 300015
- ^ a b Rodriguez IR, Mazuruk K, Schoen TJ, Chader GJ (December 1994). "Structural analysis of the human hydroxyindole-O-methyltransferase gene. Presence of two distinct promoters". The Journal of Biological Chemistry. 269 (50): 31969–31977. doi:10.1016/S0021-9258(18)31790-3. PMID 7989373.
- ^ Donohue SJ, Roseboom PH, Illnerova H, Weller JL, Klein DC (October 1993). "Human hydroxyindole-O-methyltransferase: presence of LINE-1 fragment in a cDNA clone and pineal mRNA". DNA and Cell Biology. 12 (8): 715–727. doi:10.1089/dna.1993.12.715. PMID 8397829.
- ^ Sun Q, Huang M, Wei Y (March 2021). "Diversity of the reaction mechanisms of SAM-dependent enzymes". Acta Pharmaceutica Sinica. B. 11 (3): 632–650. doi:10.1016/j.apsb.2020.08.011. PMC 7982431. PMID 33777672.
- ^ Botros HG, Legrand P, Pagan C, Bondet V, Weber P, Ben-Abdallah M, et al. (January 2013). "Crystal structure and functional mapping of human ASMT, the last enzyme of the melatonin synthesis pathway". Journal of Pineal Research. 54 (1): 46–57. doi:10.1111/j.1600-079x.2012.01020.x. PMID 22775292. S2CID 205836404.
- ^ Caspi R (2013-05-22). "Pathway: serotonin and melatonin biosynthesis I". MetaCyc Metabolic Pathway Database. Retrieved 2025-11-10.
- ^ Maltsev N, Glass E, Sulakhe D, Rodriguez A, Syed MH, Bompada T, et al. (January 2006). "PUMA2--grid-based high-throughput analysis of genomes and metabolic pathways". Nucleic Acids Research. 34 (Database issue): D369 – D372. doi:10.1093/nar/gkj095. PMC 1347457. PMID 16381888.
- ^ Fèvre-Montange M, Champier J, Szathmari A, Wierinckx A, Mottolese C, Guyotat J, et al. (July 2006). "Microarray analysis reveals differential gene expression patterns in tumors of the pineal region". Journal of Neuropathology and Experimental Neurology. 65 (7): 675–684. doi:10.1097/01.jnen.0000225907.90052.e3. PMID 16825954.
- ^ Srinivasan V, Smits M, Spence W, Lowe AD, Kayumov L, Pandi-Perumal SR, et al. (2006). "Melatonin in mood disorders". The World Journal of Biological Psychiatry. 7 (3): 138–151. doi:10.1080/15622970600571822. PMID 16861139. S2CID 21794734.
- ^ "Genetic studies probe sleep hormone's role in autism". 13 November 2011.
- ^ Ji Q, Li SJ, Zhao JB, Xiong Y, Du XH, Wang CX, et al. (May 2023). "Genetic and neural mechanisms of sleep disorders in children with autism spectrum disorder: a review". Frontiers in Psychiatry. 14 1079683. doi:10.3389/fpsyt.2023.1079683. PMC 10185750. PMID 37200906.
- ^ Yi H, Donohue SJ, Klein DC, McBride OW (February 1993). "Localization of the hydroxyindole-O-methyltransferase gene to the pseudoautosomal region: implications for mapping of psychiatric disorders". Human Molecular Genetics. 2 (2): 127–131. doi:10.1093/hmg/2.2.127. PMID 8098975.
- ^ Djeridane Y, Touitou Y (April 2001). "Chronic diazepam administration differentially affects melatonin synthesis in rat pineal and Harderian glands". Psychopharmacology. 154 (4): 403–407. doi:10.1007/s002130000631. PMID 11349394. S2CID 22918068.
- ^ Reiter RJ, Tan DX, Terron MP, Flores LJ, Czarnocki Z (2007). "Melatonin and its metabolites: new findings regarding their production and their radical scavenging actions". Acta Biochimica Polonica. 54 (1): 1–9. doi:10.18388/abp.2007_3264. PMID 17351668.
Further reading
[edit]- Itoh MT, Hosaka T, Mimuro T, Hamada N, Ishizuka B (March 2003). "Gonadotropin-releasing hormone increases melatonin release in the pineal gland of the female rat in vitro". Hormone and Metabolic Research = Hormon- Und Stoffwechselforschung = Hormones Et Metabolisme. 35 (3): 153–157. doi:10.1055/s-2003-39076. PMID 12734775. S2CID 22082149.
- Fukuda T, Akiyama N, Ikegami M, Takahashi H, Sasaki A, Oka H, et al. (May 2010). "Expression of hydroxyindole-O-methyltransferase enzyme in the human central nervous system and in pineal parenchymal cell tumors". Journal of Neuropathology and Experimental Neurology. 69 (5): 498–510. doi:10.1097/NEN.0b013e3181db7d3c. PMID 20418777.
- Donohue SJ, Roseboom PH, Illnerova H, Weller JL, Klein DC (October 1993). "Human hydroxyindole-O-methyltransferase: presence of LINE-1 fragment in a cDNA clone and pineal mRNA". DNA and Cell Biology. 12 (8): 715–727. doi:10.1089/dna.1993.12.715. PMID 8397829.
- Toma C, Rossi M, Sousa I, Blasi F, Bacchelli E, Alen R, et al. (November 2007). "Is ASMT a susceptibility gene for autism spectrum disorders? A replication study in European populations". Molecular Psychiatry. 12 (11): 977–979. doi:10.1038/sj.mp.4002069. PMID 17957233. S2CID 20794666.
- Jonsson L, Ljunggren E, Bremer A, Pedersen C, Landén M, Thuresson K, et al. (April 2010). "Mutation screening of melatonin-related genes in patients with autism spectrum disorders". BMC Medical Genomics. 3 10. doi:10.1186/1755-8794-3-10. PMC 3020629. PMID 20377855.
- Holt R, Barnby G, Maestrini E, Bacchelli E, Brocklebank D, Sousa I, et al. (September 2010). "Linkage and candidate gene studies of autism spectrum disorders in European populations". European Journal of Human Genetics. 18 (9): 1013–1019. doi:10.1038/ejhg.2010.69. PMC 2987412. PMID 20442744.
- Gałecki P, Szemraj J, Bartosz G, Bieńkiewicz M, Gałecka E, Florkowski A, et al. (May 2010). "Single-nucleotide polymorphisms and mRNA expression for melatonin synthesis rate-limiting enzyme in recurrent depressive disorder". Journal of Pineal Research. 48 (4): 311–317. doi:10.1111/j.1600-079X.2010.00754.x. PMID 20433639. S2CID 24963323.
- Yi H, Donohue SJ, Klein DC, McBride OW (February 1993). "Localization of the hydroxyindole-O-methyltransferase gene to the pseudoautosomal region: implications for mapping of psychiatric disorders". Human Molecular Genetics. 2 (2): 127–131. doi:10.1093/hmg/2.2.127. PMID 8098975.
- Sato T, Deguchi T, Ichikawa T, Fujieda H, Wake K (March 1991). "Localization of hydroxyindole O-methyltransferase-synthesizing cells in bovine epithalamus: immunocytochemistry and in-situ hybridization". Cell and Tissue Research. 263 (3): 413–418. doi:10.1007/BF00327275. PMID 1878930. S2CID 7189534.
- Rodriguez IR, Mazuruk K, Schoen TJ, Chader GJ (December 1994). "Structural analysis of the human hydroxyindole-O-methyltransferase gene. Presence of two distinct promoters". The Journal of Biological Chemistry. 269 (50): 31969–31977. doi:10.1016/S0021-9258(18)31790-3. PMID 7989373.
- Stefulj J, Hörtner M, Ghosh M, Schauenstein K, Rinner I, Wölfler A, et al. (May 2001). "Gene expression of the key enzymes of melatonin synthesis in extrapineal tissues of the rat". Journal of Pineal Research. 30 (4): 243–247. doi:10.1034/j.1600-079X.2001.300408.x. PMID 11339514. S2CID 5800718.
- Melke J, Goubran Botros H, Chaste P, Betancur C, Nygren G, Anckarsäter H, et al. (January 2008). "Abnormal melatonin synthesis in autism spectrum disorders". Molecular Psychiatry. 13 (1): 90–98. doi:10.1038/sj.mp.4002016. PMC 2199264. PMID 17505466.
External links
[edit]- Acetylserotonin+N-Methyltransferase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- ASMTL+protein,+human at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Human ASMT genome location and ASMT gene details page in the UCSC Genome Browser.